Methods
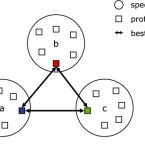
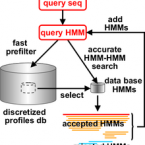
VOG are created from scratch for all viral genomes from NCBI RefSeq. VOG are built fully automated, based on manually curated reference data (annotation quality criteria) and associations (functional categories). Our VOG workflow is controlled by Python scripts and is mainly based on NCBI COGsoft and HH-suite (see below).